High-throughput cell DNA screening using digital PCR
High-throughput screening is one of the main challenges in current scientific studies, as it is a powerful tool to detect the presence of specific mutations or activities in complex cell populations. To acquire a more detailed understanding of cellular expression at the single-cell level, compartmentalization of biological compounds inside water-in-oil droplets using microfluidic technologies is gaining popularity. With controlled DNA molecule encapsulation using droplet microfluidics, Michael Ryckelynck and his team are able to analyze enzyme production resulting from DNA expression.
See the Application Note made in collaboration with Michael Ryckelynck et al., ISIS Team, University of Strasbourg
Introduction to DNA Screening Using Digital PCR
Polymerase Chain Reaction, or PCR, has an almost ubiquitous presence in biomedical sciences. Due to the ease of execution and the exquisite sensitivity offered by PCR, which covers amplification from a single template molecule up to about nine orders of magnitude, the applications of PCR are widespread. This includes targeted mutagenesis, DNA screening using digital PCR, virus diagnosis and analysis, and more. (1).
Droplet microfluidics offers significant advantages for performing high-throughput screens and sensitive assays. Droplets make it possible to considerably reduce sample volumes, which lowers costs. Compartmentalization in droplets increases assay sensitivity by increasing the effective concentration of rare species and decreasing the time required to reach detection thresholds. Droplet microfluidics combines these powerful features to enable previously inaccessible high-throughput screening applications, including single-cell and single-molecule assays (2).
Droplet digital PCR (ddPCR) is based on the amplification of single target DNA molecules in many separate droplets, which provides a new method for accurate quantification of DNA copy numbers. In a ddPCR assay, the distribution of target DNA molecules among the reactions follow Poisson statistics, which means that the majority of reactions contain either one or zero target DNA molecules. DdPCR has several advantages over more traditional qPCR methods.
- It enables the absolute quantification of target nucleic acid without the reliance on rate-based measurements and the need to use calibration curves.
- It demonstrates high sensitivity and precision for low-copy-number target nucleic acids. (2,3).
In this application note, the objective is to use Fluigent products to perform DNA screenings using digital PCR by isolating individual DNA molecules and analyzing the enzymes resulting from their expression.
How to perform high-throughput digital DNA screening
Materials: Products
Fluigent pressure-driven flow control solutions are ideal for high-throughput droplet generation for DNA screening, as the excellent flow stability offered by this technology ensures high droplet monodispersity, a controlled encapsulation rate, and experimental reproducibility.
Methods
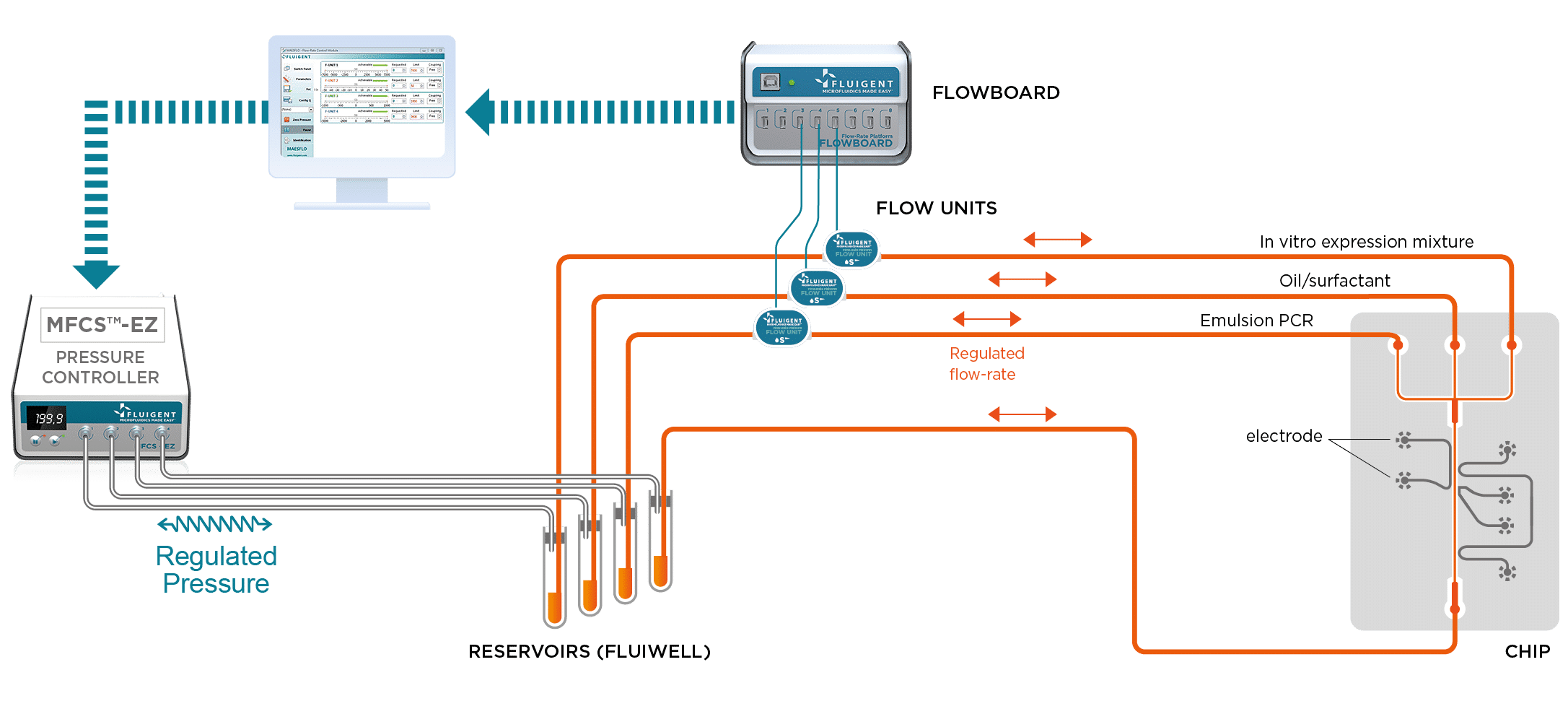
For these studies, it is important that the droplets be generated at the correct frequency and at a uniform size. Using the MFCS™-EZ in this experiment is critical, as the device generates stable flows and precise control of the different phases.
Part one: generation of the pcr/dna droplets
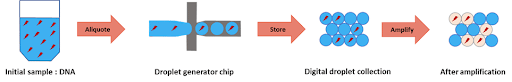
To perform DNA high-throughput digital screening using digital PCR, two separate emulsions were first generated. The first one encapsulated an aqueous phase of a PCR and DNA mixture in an organic continuous phase on a dedicated chip. To do this, the initial sample is injected into the microfluidic droplet generator, where it is cut into highly-monodisperse droplets by a perpendicular stream of perfluorinated oil (figure 2). Each droplet contains either one or zero molecules of DNA. The amplification step triggers the activation or production of a fluorescent dye in the droplets containing the targeted DNA. Counting the number of fluorescent droplets is equivalent to counting the number of DNA molecules.
Part two: fusion of one pcr droplet with one ivt droplet generated on a chip
A green fluorescent marker is added to the PCR primers to make them visible at the end of the process. These droplets either contain DNA to be enhanced or are empty. An orange dye is also added to measure the size of the droplet, where the intensity of the light captured by a CCD camera is proportional to the size of the droplet.
For DNA screenings using digital PCR, after amplification, the PCR droplets must be injected into a second chip. This chip generates IVT droplets, while also synchronizing them to create one PCR droplet that is inserted between two IVT droplets. The IVT droplets contain an orange dye to differentiate the contents. Fusion is processed by the application of an electric field.
Results
Our set-up features fast start-up and an automated configuration that allows the experiment to be performed without constant adjustments, thus saving time and enabling systemic screening. By continuously measuring the lows and pressures, the performance of the fluidic system can be evaluated at a glance, providing full control of the process.
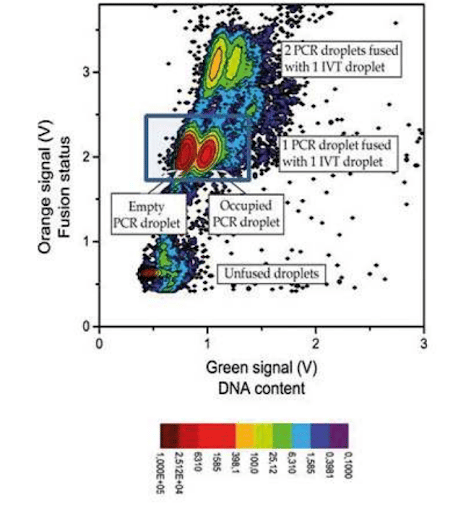
In our experimental DNA screening using digital PCR, we obtained a 1/1 fusion at an 85% to 88% success rate (no fusion: 10%, double fusion: 5%) with stable and synchronized decays.
This graph represents a count of droplets with given orange and green signals. The darker red dot signifies a large number of droplets with these characteristics, while light blue represents a low number of droplets. The green signal shows the presence of DNA in the PCR droplet, and the orange one is relevant for detecting the occurrence of fusion.
Conclusion
To perform DNA screening using digital PCR, it is essential to have high stability in order to obtain reproducible and optimal results. Flow-rate control solutions based on pressure actuation provide:
- High droplet monodispersity (even at low low-rates over long time periods.)
- Straightforward, easily automated setup
- Precise volume and low control – even with complex, multi-channel chips to minimize cross-talk between low channels.
- Ability to detect and compensate for small disruptions such as air bubbles.
Related resources
References
- Hoshino, T. and Inagaki, F. (2012) “Molecular quantification of environmental DNA using microfluidics and digital PCR,” Systematic and Applied Microbiology, 35(6), pp. 390–395. Available at: https://doi.org/10.1016/j.syapm.2012.06.006.
- Payne, E.M. et al. (2020) “High-throughput screening by droplet microfluidics: Perspective into key challenges and future prospects,” Lab on a Chip, 20(13), pp. 2247–2262. Available at: https://doi.org/10.1039/d0lc00347f.
- Markey, A.L., Mohr, S. and Day, P.J.R. (2010) “High-throughput droplet PCR,” Methods, 50(4), pp. 277–281. Available at: https://doi.org/10.1016/j.ymeth.2010.01.030.