Development of a human gut-on-chip to assess the effect of shear stress on intestinal functions
Fluid flow and shear stress have previously been reported to promote proper intestinal cell differentiation, formation of villus-like 3D structures and enhanced intestinal barrier function.
Our gut-on-chip system made of BEOnChip microfluidic chips coupled to pressure controllers offers a way to control and mimic the microenvironment of in vitro cultured intestinal epithelium that is closer to the physiological state.
BEOnChip offers disposable systems to recreate in-vitro biological enviroments using microtechnologies.

Introduction to Gut-on-Chip Technology
Why Study Intestine-On-Chip?
The intestine stands as a complex and pivotal organ, orchestrating the absorption of nutrients and water while playing a vital role in immunological functions. Serving as the primary conduit for drug absorption, it also hosts a diverse microbiota crucial for digestion and nutrient absorption. Dysfunctions in the intestine can lead to severe and chronic gut-related disorders, including Inflammatory Bowel Disease (IBD) such as ulcerative colitis, Crohn’s disease, celiac disease, and Irritable Bowel Syndrome.
The global rise in the incidence of intestinal disorders, particularly IBDs, underscores the need for a deeper understanding of their origins. The etiology of these disorders remains incompletely understood, with a complex interplay between the host immune system, genetics, microbiota, and environmental factors being the most widely acknowledged contributing factors.
The imbalance between pro-inflammatory and anti-inflammatory cytokines, along with alterations in the composition and function of the gut microbiota (dysbiosis), plays a pivotal role in these conditions.
Traditionally, research on the gut microbiome relies heavily on laboratory mice. However, these animal models often fall short when extrapolated to human contexts due to differences in microbiota composition and the immune system. Enter Gut-on-Chip technology—an innovative approach that holds promise for more accurate and relevant investigations into intestinal functions and disorders, bridging the gap between traditional models and human physiology.
Organ-on-Chip Advancements for Gut Studies
In recent years, the revolutionary field of organ-on-chip (OOC) technology has emerged as a highly promising solution to address the limitations of animal models. This innovative approach involves replicating tissue and organ-level physiology within biologically inspired microfluidic in vitro devices. Unlike traditional 2D monoculture plates, OOC devices, particularly gut-on-a-chip (GOC) systems, recreate the intricate microenvironment found in vivo, providing a more accurate representation of gut dynamics and host-microbiome interactions.
Gut-on-chip technology specifically aims to mimic the structure, function, and microenvironment of the human gut. It has proven to be a powerful tool for constructing physiological models of the human gut, facilitating drug testing and development, and exploring host–microbiome interactions (1). The BE-FLOW microfluidic chip, crafted from cyclic olefin polymer (COP), serves as an impermeable material for gases, enabling precise control over gas concentrations within the devices.
Furthermore, recognizing the influence of peristalsis on the differentiation of cells lining the digestive tract, devices that induce fluid flow and shear stress have been instrumental. These devices promote the accelerated differentiation of intestinal epithelial cells, foster the formation of three-dimensional villus-like structures, and enhance intestinal barrier function. This innovative technology effectively replicates the intricate functions of the human intestine, advancing our understanding of intestinal physiology and disease etiology.
What are the Advantages of this Approach?
In contrast to traditional model systems, gut-on-chip technology provides a multitude of benefits, including real-time observation, simulation of the intestinal microenvironment, adjustable fluid flow shear stress (1), and a dynamic host–microbiome interface (1).
To establish a comprehensive model of the human gut-on-a-chip, we introduce a complete system that integrates BE-FLOW chips with Fluigent pressure-based flow controllers. This innovative setup offers precise control over the microenvironment of the intestinal epithelium, aligning more closely with its physiological state.
Method for Developing a Human Intestine-On-Chip
Selecting the Appropriate Microfluidic Chip to Mimic Intestinal Functions
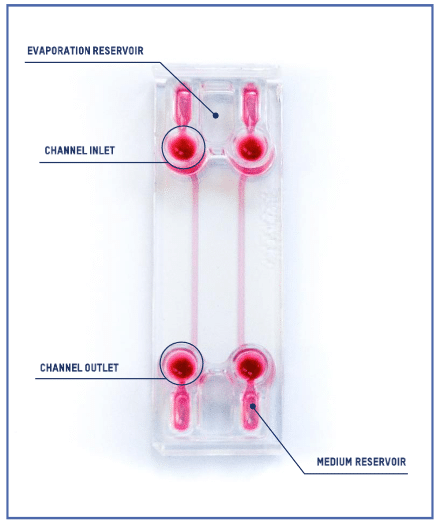
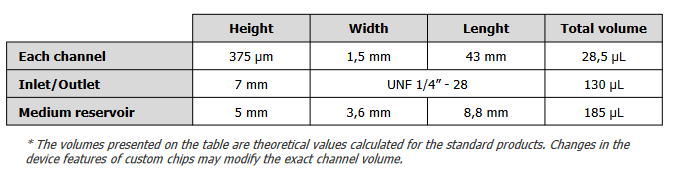
Introducing the BE-FLOW Gut-on-Chip Device
The BE-FLOW microfluidic chip is designed with two independent channels featuring threaded inlets and outlets, allowing the seamless integration of connectors and tubing connected to fluidic controllers.
This design facilitates the application of an independent flow rate in both channels through any perfusion system. To prevent culture media evaporation at the beginning of the experiment (prior to perfusion initiation), water-filled reservoirs are strategically placed adjacent to the medium reservoir (refer to the scheme for details).
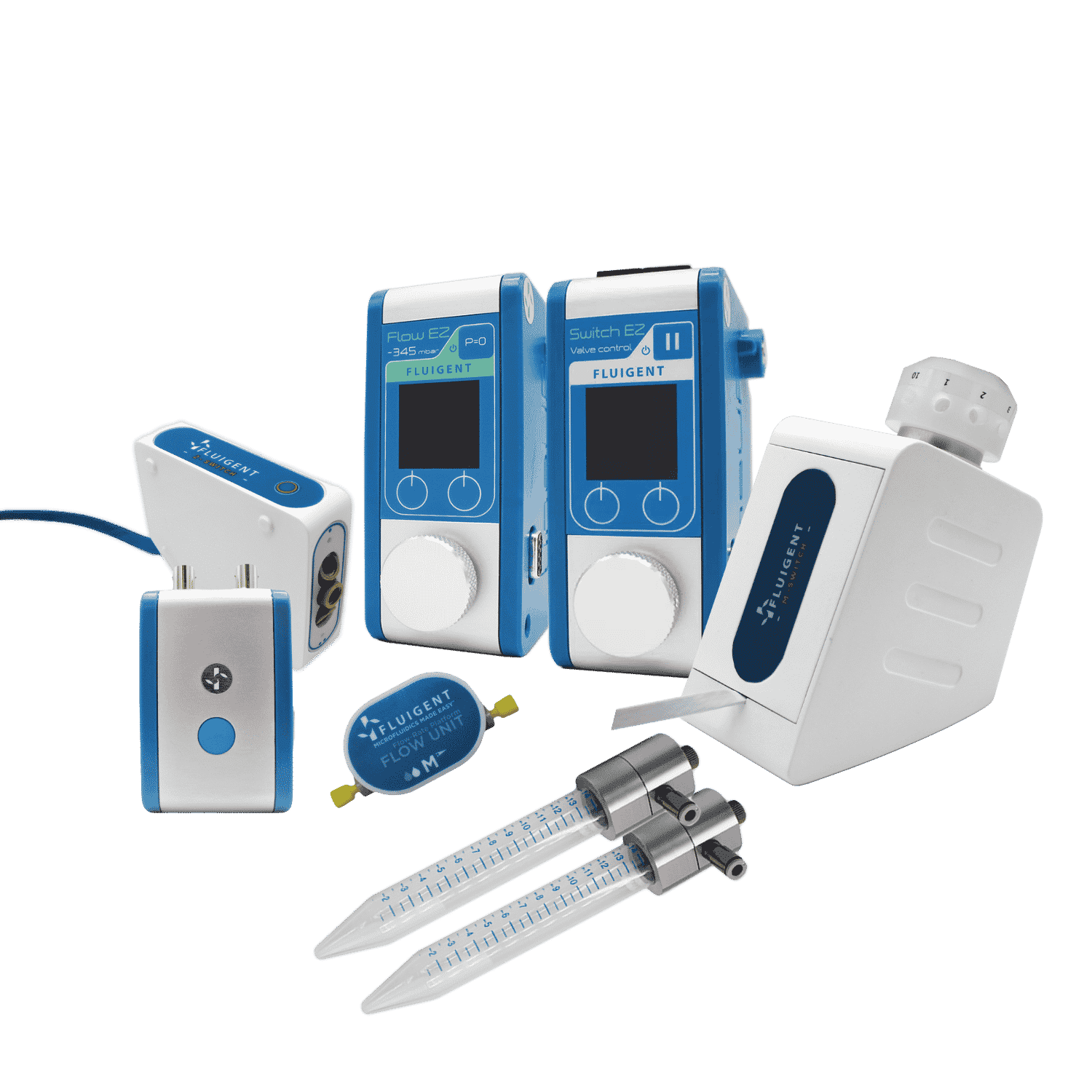
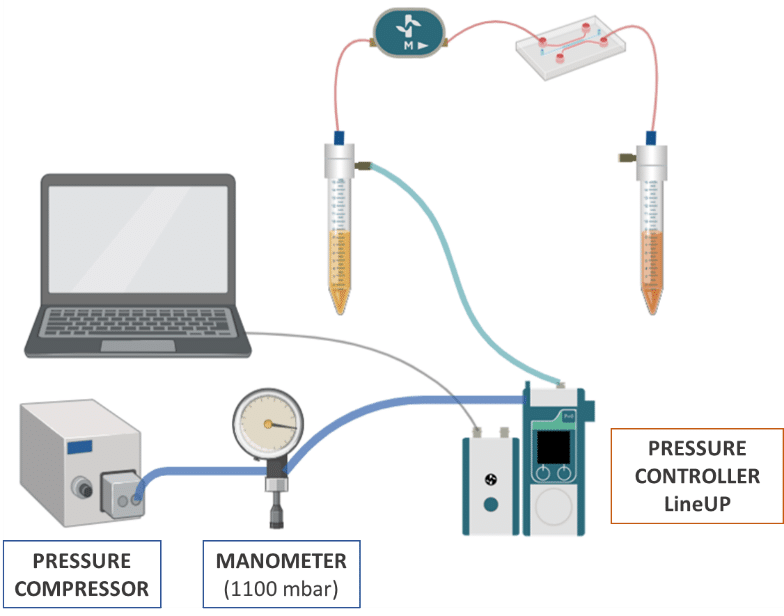
Pressure and Flow Control Instruments for Gut-on-Chip Experiments
Fluigent positive pressure controllers utilize low air pressures (mbar) to compel a solution to ascend through an output tube, ensuring precise control over the flow rate in a microfluidic system.
The configuration of the fluidic system is illustrated in the illustration:
- Pressure compressor RX
- Regulator
- FlowEZ (1000 mbar)
- FlowUnit S
How to perform Gut-on-a-Chip development?
BE-FLOW cell culture
Before seeding, pre-warm the Be-Flow standard device in the incubator overnight to minimize the formation of air bubbles.
- Fill the channel with 100μl of collagen at 0.1 mg/ml (in PBS 1X) and incubate at 37°C for 30min. Wash the channel by adding 100μl of PBS 1X into the inlet and by aspirating it at the outlet using P200 pipette. Repeat this step three times.
- Aspirate PBS 1X completely before seeding.
- Seed 1.106 Caco-2 cells resuspended in 50μl of culture media.
- Incubate at 37°C, 5% CO2 until the cells have properly spread (2-4h).
- Once cell spreading has occurred, add 300μl of culture medium into the medium reservoirs. Add H2O to the evaporation reservoirs and cover.
If connection to a perfusion system is not possible for more than 2 hours following cell seeding, keep the device in the incubator and renew the culture medium using a rocker as long as needed.
Microfluidic intestine-on-chip Set Up
Before setting the flow up:
- Sterilize and pre-warm the tubes and the fluidic elements overnight at 37°C.
- Set the system in a laminar flow cabinet.
- The channels and inlet/outlet wells should never be depleted of culture medium.
- Both inlets and outlets are designed to be able to use connectors (1/4’’- 28).
To work with cells, the assembly of the circuit must be carried out under sterile conditions under a biosafety cabinet. Before use, autoclave the CAPs, pneumatic tubes, threaded connections and ferrules to be used.
- Connect all circuit components except the microfluidic device.
- Establish a flow of ethanol (70%) for at least 15 min to sterilize the sensor and then wash with abundant sterile H2O. Dry by passing air through at maximum pressure. The sensor CANNOT be autoclaved.
- Prime the tubes of the circuit with culture medium until there are no air bubbles
- Remove the culture medium from the chip reservoirs (not from inlet/outlet wells) and connect the tubes to the outlet/inlet using the threaded connections and ferrules.
- The ferrules should be manipulated with the help of sterile forceps.
- Ensure that the tube is perfectly fixed.
- Remove the displaced medium from the reservoirs.
Once the system is closed, switch the flow on at 5.8µl/min (shear stress 0.02 dyn/cm²) for 4 days to promote villi formation.o Observe the system under perfusion for a few minutes to check that there are no leaks.
Results: Increase In Cell Density
Upon completing 4 days of continuous perfusion, the flow is halted, and the gut-on-chip system is positioned under an inverted microscope for the observation of cell behavior and differentiation. As depicted in Figure 1, there is a noticeable increase in cell density following 4 days of culture under dynamic flow conditions. Additionally, Caco-2 cells exhibit the initiation of 3D structure formation, a feature not observed under static conditions.
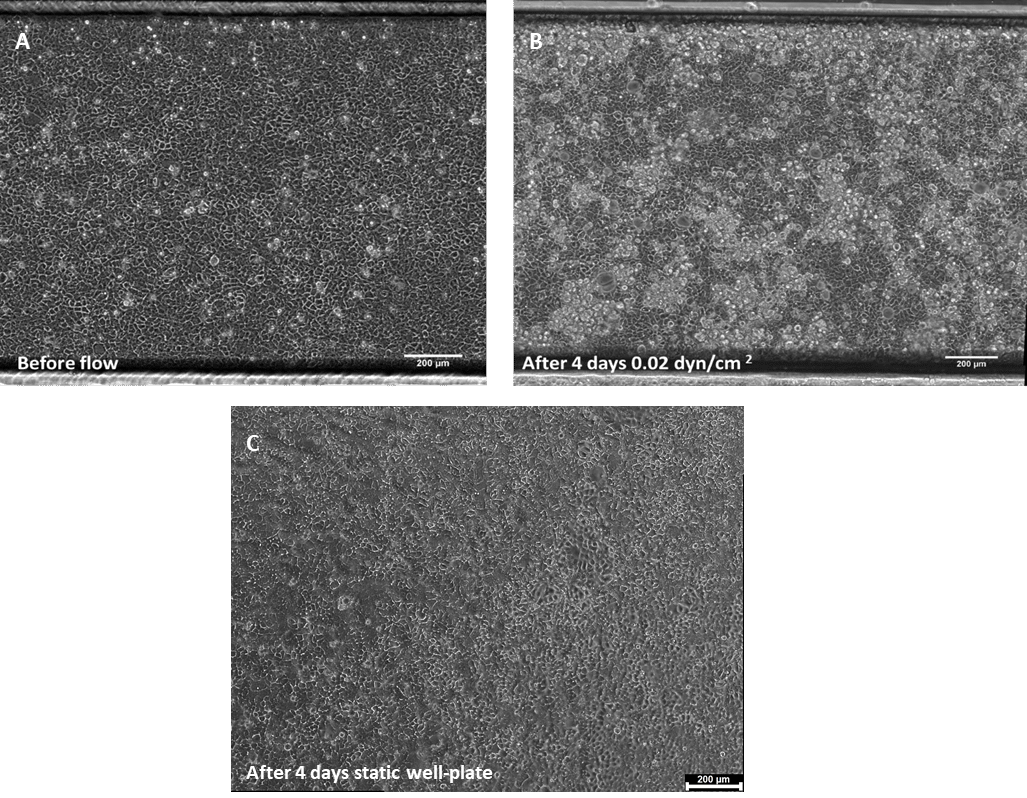
Figure 1. Phase contrast images of Caco-2 monolayer in a BE-FLOW channel (A) 24h post cell seeding
before the addition of flow, and after 4 days under shear stress conditions (B), or static conditions
(C). Scale bar = 200 μm.
References
(1). Ashammakhi, N., Nasiri, R., Barros, N., Tebon, P., Thakor, J., & Goudie, M. et al. (2020). Gut-on-a-chip: Current progress and future opportunities. Biomaterials, 255, 120196. doi: 10.1016/j.biomaterials.2020.120196
Related Resources
- Expert Reviews: Basics of Microfluidics
How to reproduce active biomimetic stimulation in vitro?
Discover - Microfluidics White Papers
A review of Organ on Chip Technology – A White Paper
Discover 
White paper: Organ on Chip
Discover- Interviews & Testimonials
Panel Discussion & Interviews – Microfluidics & Organ-On-Chips
Discover 
WEBINAR: An one-of-a-kind Organ-on-chip platform
Discover