Mikrofluidik für die Tröpfchenerzeugung
Was sind Emulsionen und Tröpfchen?
Eine Emulsion ist ein heterogenes System, das aus mindestens einer nicht mischbaren Flüssigkeit besteht, die in einer anderen in Form von Tröpfchen dispergiert ist. Wenn nicht emulgiert wird, werden die beiden Flüssigkeiten getrennt, wobei die weniger dichte Phase auf der dichteren Phase liegt. Salatdressings sind gängige Beispiele für Emulsionen. In einer Vinaigrette, die aus einer wässrigen Phase (dem Essig) und einer öligen Phase (Olivenöl) besteht, werden die beiden Phasen getrennt, wenn sie nicht emulgiert werden. Durch Schütteln der Vinaigretteflasche wird der Essig in der kontinuierlichen Ölphase verteilt, so dass eine Emulsion entsteht. Bei einer Emulsion wird eine Flüssigkeit (die sogenannte dispergierte Phase) in der anderen (der kontinuierlichen Phase) dispergiert.
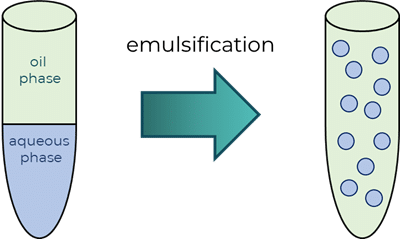
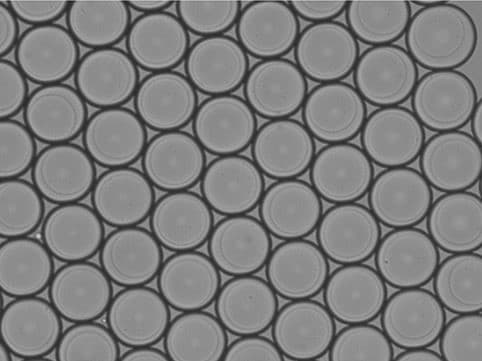
Wasser in Öltröpfchen
Was sind die häufigsten Anwendungen von Emulsionen?
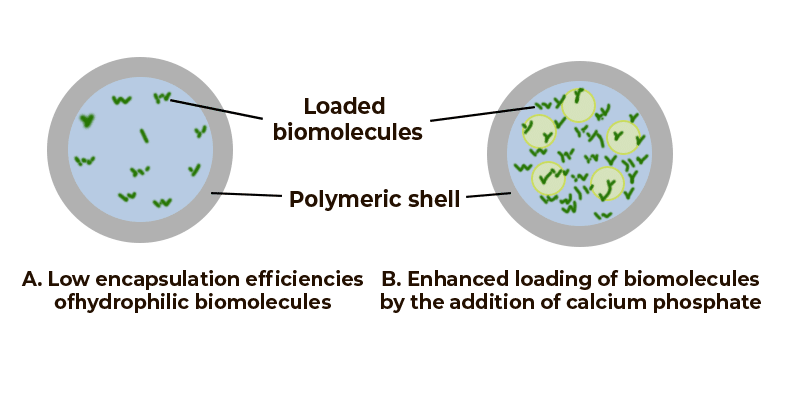
Verkapselung von Biomolekülen in Polymeren [1]
Pharmazie: Emulsionen werden für eine Vielzahl pharmazeutischer Produkte verwendet, darunter intravenöse, intramuskuläre, okuläre oder orale Produkte. Emulsionen werden auch als Vorlagen für Polymer-Mikropartikel, Lipid-Nanopartikel oder Mikrokapseln verwendet. Letztere können z. B. für die Verabreichung von Arzneimitteln verwendet werden, wobei die Emulsion selbst der aktive pharmazeutische Wirkstoff (API) oder ein Adjuvans für die gleichzeitige Verabreichung ist.
Lebensmittelindustrie: Durch die einzigartige Natur von Emulsionen als thermodynamisch stabile Dispersionen, die mit einer Größenverteilung unterschiedlich großer Tröpfchen erzeugt werden, sind sie für verschiedene Anwendungen in der Lebensmittelindustrie geeignet.
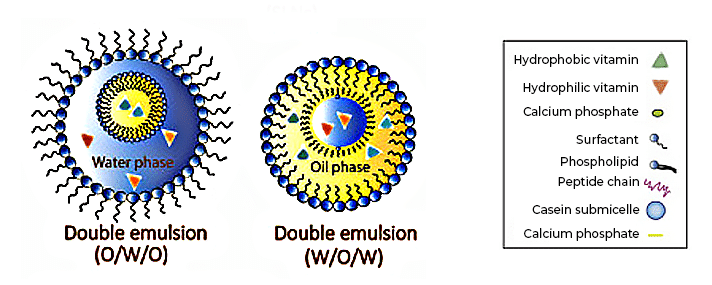
Die Nanoencapsulation, in diesem Beispiel ein vielversprechender Ansatz für die gezielte Abgabe und kontrollierte Freisetzung von Vitaminen. [2]

Kosmetische Produkte, die mit Hilfe der Mikrofluidik-Technologie entwickelt wurden.
Kosmetik: Produkte auf Emulsionsbasis in Kosmetik bieten eine feine Textur und ein angenehmes Gefühl beim Auftragen. Sie können auch die Löslichkeit von relativ schwach löslichen Stoffen verbessern. In letzter Zeit sind „Millifluidik“-Geräte auf den Markt gekommen, mit denen Emulsionen erzeugt werden können, die mit bloßem Auge sichtbar sind, was die Entwicklung einzigartiger Produkte mit hoher optischer Attraktivität ermöglicht.
Warum die Tropfen-mikrofluidik?
1) Die Grenzen der herkömmlichen Methoden zur Tropfenerzeugung
Zu den Standardmethoden für die Tropfenerzeugung gehören mechanische Geräte wie Hochgeschwindigkeitsmischer, Hochdruckhomogenisatoren und Kolloidmühlen.
Das Aufbrechen von Tropfen erfolgt in der Regel durch Scher- oder Stoßbelastungen, die durch manuelles/mechanisches Rühren erzeugt werden. Unter solchen Bedingungen sind die erzeugten Spannungen in der Regel nicht gleichmäßig über das System verteilt. Infolgedessen haben die erzeugten Emulsionen eine polydisperse Größe. Dies kann bei vielen Anwendungen eine starke Einschränkung darstellen, da die Stabilität von Emulsionen von der Größe abhängt.
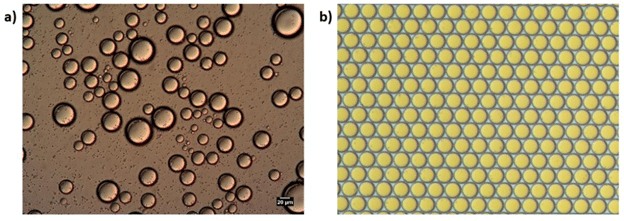
Herstellung von Mikroemulsionen mittels a) Batch-Verfahren und b) mikrofluidischem Verfahren
2) Mikrofluidische Geräte für kontrollierte Emulsionen
Die Tropfenerzeugung mit mikrofluidischen Systemen wurde für Anwendungen eingeführt, bei denen die Monodispersität von großer Bedeutung ist. In mikrometergroßen Kanälen wird jeweils ein Tropfen erzeugt, was die Herstellung monodisperser Tropfen ermöglicht. Mit einem solchen Maß an Kontrolle sind Anwendungen möglich geworden, die vorher nicht möglich gewesen wären, wie z. B. die digitale PCR und die Verkapselung einzelner Zellen in Tröpfchen. Es ist auch eine hervorragende Methode für Anwendungen, die teure API (Active Pharmaceutical Ingredient) verwenden, da weniger Abfall anfällt. In einem typischen mikrofluidischen System ist der mikrofluidische Chip mit einem oder mehreren Fluidikreglern verbunden, die die Flüssigkeiten in den Chip injizieren. Die Tropfenerzeugung hängt hauptsächlich von der Größe der mikrofluidischen Kanäle, den Flüssigkeitseigenschaften (Viskosität) und den verwendeten Durchflussraten ab.
Mikrofluidik-basierte Tropfenerzeugung und -kontrolle ermöglicht :
- Hochgradig monodisperse (<2% Größenvariation) Tropfenproduktion, im Gegensatz zu Batch-Emulsionsmethoden, mit relativ hoher Frequenz
- Hochgradig reproduzierbare komplexe Strukturen (Mehrfachemulsionen, Emulsionen mit mehreren Kernen …)
- Manipulation eines einzelnen Tropfen als individueller biochemischer Reaktor im pL-Maßstab.
- Miniaturisierung von Produktions- und bioanalytischen Geräten
Überwindung der Beschränkungen bei den Materialien und der Oberflächenbehandlung von Dropletchips.
Die meisten handelsüblichen mikrofluidischen Tropfengeneratoren basieren auf einer planaren, strömungsfokussierenden Konfiguration, die in Polymer- oder Glas-Chips implementiert ist. Diese Geometrie hat viele Einschränkungen, wie die Notwendigkeit spezifischer Beschichtungen oder die Verwendung spezieller Tenside.
Im Gegensatz dazu stellen auf Glaskapillaren basierende Geräte für die Tropfenerzeugung eine große Verbesserung dar, da die dispergierte Phase nie mit den Wänden der äußeren Kapillare in Kontakt kommt (a,b).
Sie sind schwierig zu implementieren (Zentrierung der Kapillare), und kommerziell verfügbare Designs haben sich als wenig flexibel für die Tropfenproduktion erwiesen (> 100 µm Durchmesser, < 1 kHz Erzeugungsrate). Die Zentrierung kann vereinfacht werden, indem zwei kreisförmige Kapillaren in eine quadratische äußere Strömungskapillare eingesetzt werden ( c,d), aber die damit verbundenen Herstellungsverfahren schränken die Produktion in großem Maßstab ein, und eine kapillarbasierte Tropfenproduktion mit hohem Durchsatz ist noch nicht erreicht worden. Eine neue Konfiguration bietet eine vielversprechende Alternative, bei der das Extraktionsrohr vor dem Injektionsrohr platziert wird, ohne dass die Umgebung eingeschlossen wird (e). Dieses System funktioniert jedoch nur im Jetting-Regime, was nicht die Monodispersität der Tropfen garantiert, die mit dem Dripping-Regime verbunden ist.
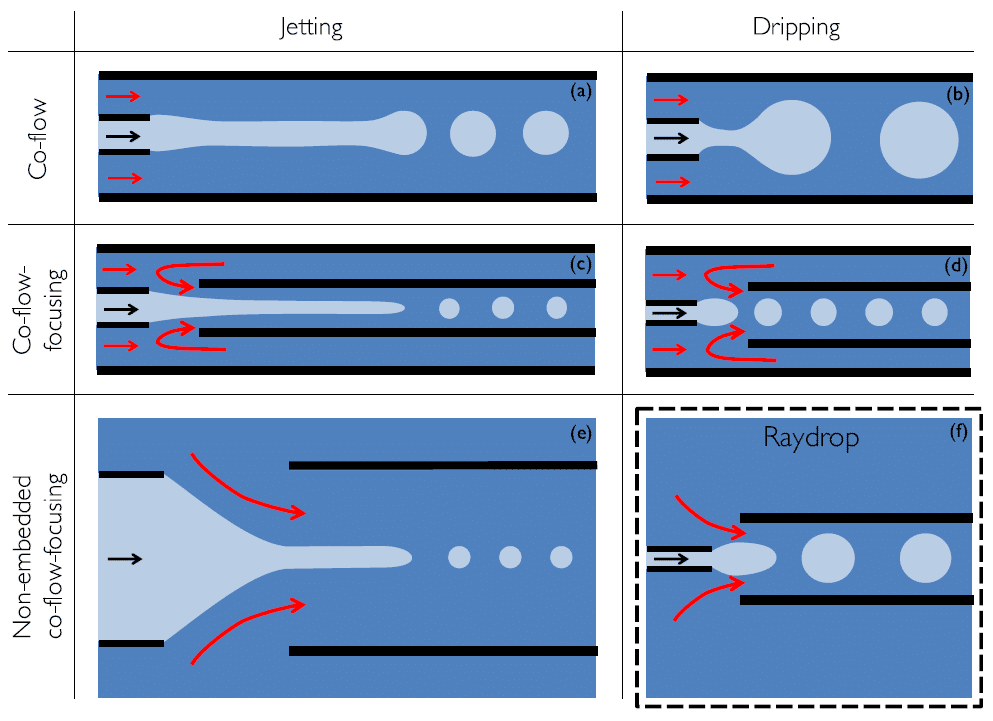
Verfügbare kapillarbasierte achsensymmetrische Designs von Tröpfchengeneratoren.
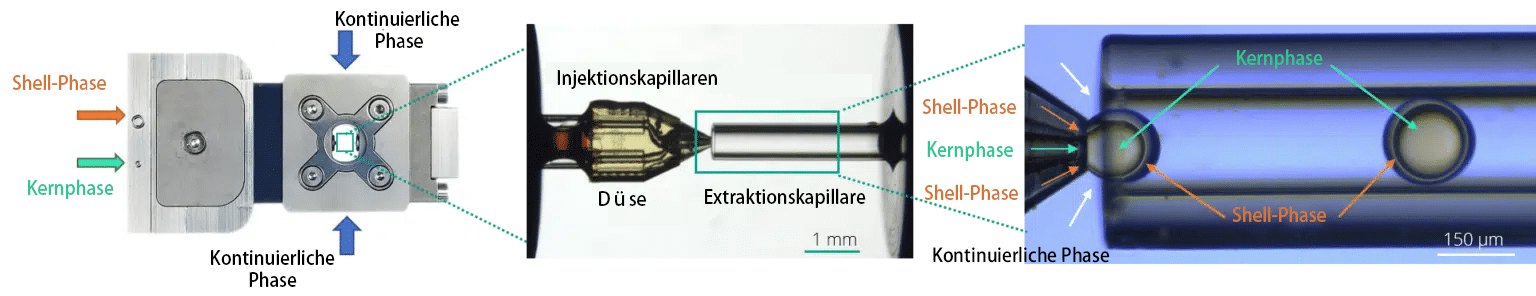
Raydrop: Ein nicht eingebetteter mikrofluidischer Tropfengenerator mit Co-Flow-Fokussierung
Fluigent und Secoya haben ihr Fachwissen gebündelt, um ein neues System zu entwickeln, das auf der letztgenannten Konfiguration basiert, bei dem jedoch das Tropfsystem durch die Verwendung einer Injektionskapillare mit einem kleineren Durchmesser als die Extraktionskapillare erzwungen wird. Erreicht wird dies durch eine verbesserte Kombination aus modernsten Bearbeitungs- und 3D-Drucktechniken. Dieses Design weist sowohl die Merkmale einer Co-Flow- als auch einer Flow-Focusing-Konfiguration auf und wird als “ Co-Flow-Focusing-Design“ bezeichnet. Diese Konfiguration füllt die Lücke in der Designpalette der mikrofluidischen Tropfengeneratoren.
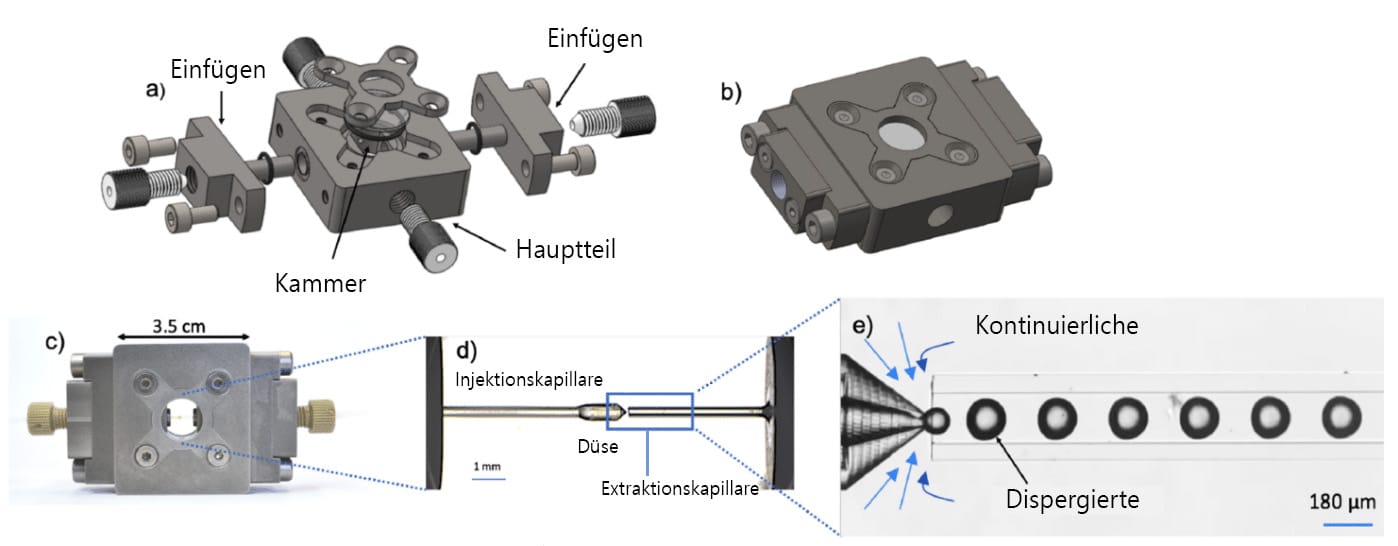
a) Explosionszeichnung und b) zusammengesetzte Ansicht des Raydrop. (c) Der Raydrop mit den Injektions- und Extraktionskapillaren aus Glas. (d) Zoom durch das obere Fenster der beiden Kapillaren, die in der mit der kontinuierlichen Phase gefüllten Kammer angeordnet sind. (e) Zoom auf den kapillarbasierten Tropfen-erzeugungsbereich.
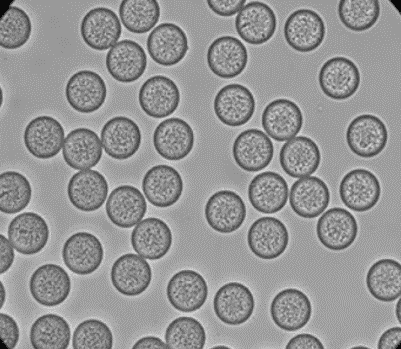
Beispiele von PLGA-Mikroperlen
Der Raydrop ist ein einfach zu bedienender, kommerziell erhältlicher mikrofluidischer Chip, der es ermöglicht, Tropfen mit hervorragender Monodispersität zu erzeugen.
Der Raydrop® ermöglicht die Herstellung von einfachen Emulsionen (Öl-in-Wasser und Wasser-in-Öl) sowie von Doppelemulsionen (Öl-in-Wasser-in-Öl oder Wasser-in-Öl-in-Wasser) ohne Beschichtung.
Doppelemulsionen haben im Vergleich zu einfachen Emulsionen viele Vorteile. Sie sind in der Regel stabiler, neigen weniger zur Koaleszenz und sind einfacher zu handhaben und zu analysieren. Da sich Doppelemulsionen leicht in einer wässrigen kontinuierlichen Phase dispergieren lassen (was bei Einfachemulsionen im Bereich der Biowissenschaften weniger der Fall ist), können sie mit automatischer Zellsortierung wie der fluoreszenzaktivierten Zellsortierung (FACS) charakterisiert und sortiert werden. Darüber hinaus eignen sich Doppelemulsionen hervorragend zur Herstellung von Mikrokapseln durch Verfestigung der Hülle.
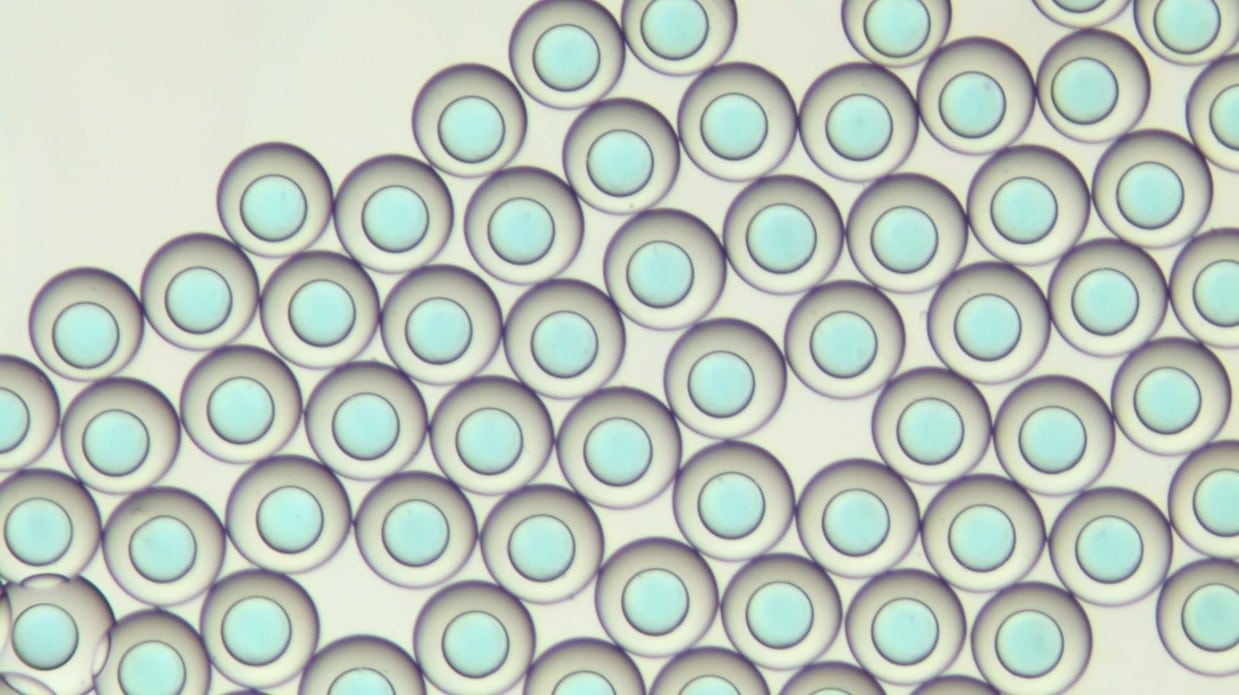
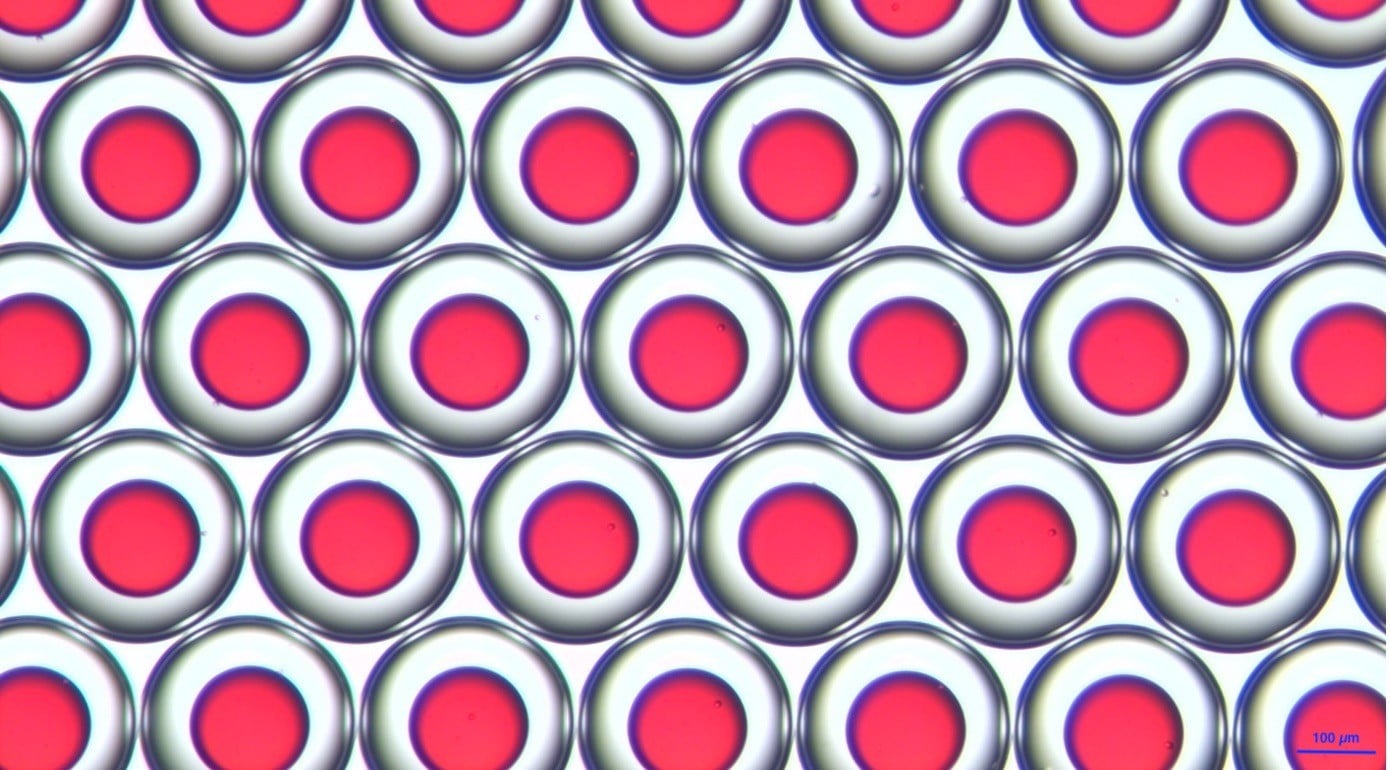
Beispiele von PLGA- (links) und Chitosan-Mikrokapseln (rechts)
Beispiele von PLGA- (links) und Chitosan-Mikrokapseln (rechts)
Welche Rolle spielt die Durchflussrate bei der Tröpfchenbildung?
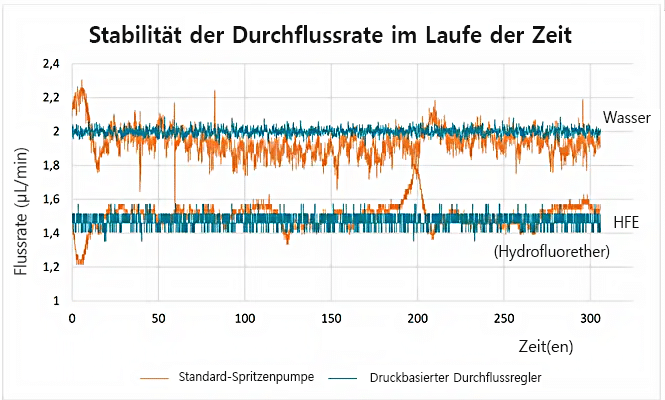
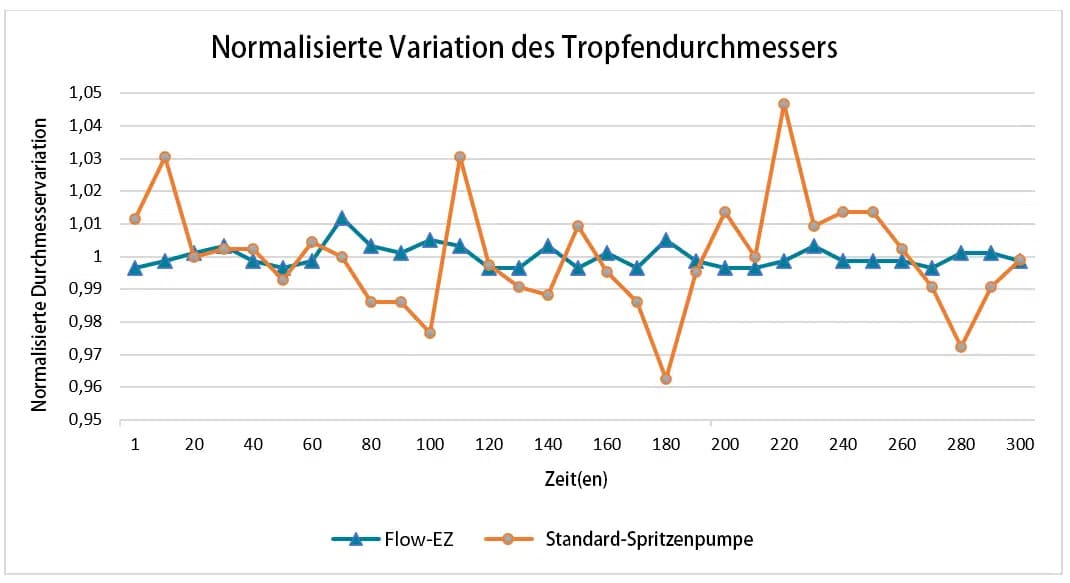
Die Stabilität der Durchflussrate ist entscheidend für wiederholbare Reaktorvolumina und reproduzierbare Ergebnisse. Spritzenpumpen werden häufig für die Erzeugung von Tröpfchen in mikrofluidischen Experimenten verwendet. Je nach Modell haben Spritzenpumpen eine begrenzte Durchflusskontrolle. Daher wird die Tröpfchengröße, die proportional zur Durchflussrate ist, beeinflusst. Die tatsächliche Durchflussrate kann mit solchen Geräten nicht überwacht werden. Der Wert der Flussrate wird auf dem Gerät angezeigt, aber es gibt keine Informationen über die Zeit, die benötigt wird, um eine bestimmte Flussrate zu erreichen (die Zeit für das Gleichgewicht der Durchflussrate kann je nach mikrofluidischem Aufbau variieren, und die Durchflussrate kann je nach Gerät schwanken). Eine Alternative zu Spritzenpumpen sind unsere druckbasierten Flow EZ-Druckregler. Diese zeigen, dass eine hochpräzise Flusskontrolle, eine hohe Reaktionszeit und eine Flussüberwachung möglich sind. Im Vergleich zur Spritzenpumpe wurde ein stabiler und wiederholbarer Fluss erzeugt.
Referenzen
- Iqbal, M., Zafar, N., Fessi, H., & Elaissari, A. (2015). Double emulsion solvent evaporation techniques used for drug encapsulation. International Journal of Pharmaceutics. https://doi.org/10.1016/j.ijpharm.2015.10.057
- Katouzian, I., Jafari, S.M., Nano-encapsulation as a promising approach for targeted delivery and controlled release of vitamins, Trends in Food Science & Technology (2016), https://doi: 10.1016/j.tifs.2016.05.002