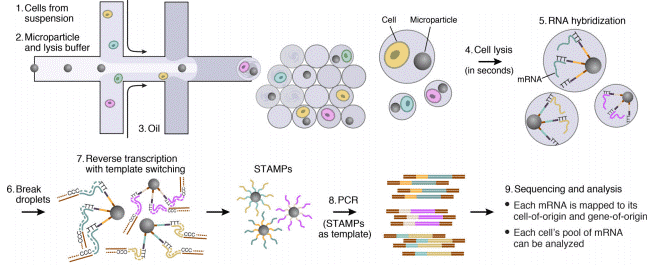
Droplet Sequencing: Drop-Seq method
The Droplet Sequencing method, known as Drop-Seq, is a method for quickly profiling thousands of individual cells by separating them into nanoliter-sized aqueous droplets, associating a different barcode with each cell’s RNAs, and sequencing them all together. The Drop-Seq method analyzes mRNA transcripts from thousands of individual cells simultaneously while remembering transcripts’ cell of origin.
The encapsulation process is carried out with a microfluidic flow-focusing junction chip (PN:ODROPSEQC) designed and developed by the company FlowJEM, which provides Drop-Seq microfluidic technology to the Macosko and McCarroll labs for their Next Generation Sequencing (NGS) experiments using the Drop-Seq protocol.
Principle of the Droplet-sequencing method
Droplet-sequencing, also known as single-cell sequencing in droplets, is a cutting-edge technique revolutionizing the field of genomics.
The Drop-Seq protocol, originally developed by Macosko et al. in 2015, is a high throughput method that enables the sequencing of the mRNA from a large number of cells. The power of this technology resides in the fact that during sequencing, one can distinguish where the original information came on a cell to cell basis. This allows one to make a gene expression map of the cell, or even to distinguish cell populations within a tissue.
This Drop-Seq method relies on droplet microfluidics and library preparation for Next-Gen Sequencing (NGS): droplets allow for rapid and efficient compartmentalization using low reagent volumes and NGS allows for fast and high throughput analysis of single-cell gene expression.
The Droplet-Sequencing method relies on pairing one barcoded bead and one cell in a small droplet. After droplet formation, cell lysis occurs such that the polyadenylated RNA produced exclusively by this cell can specifically be captured on the bead. Capturing RNA with a single barcoded bead will allow the reconstitution of the information coming from that cell.
Next, the mRNA is reverse transcribed along with the barcode into tagged cDNA, and PCR amplified. Finally, the resulting libraries are sequenced by NGS, and analyzed using various bioinformatics tools: cell barcode sequences are identified, transcript sequences linked to the barcodes mapped to a reference genome. All transcripts linked to one barcode form the gene-expression profile of a single cell
What about the drop-seq process?
All the reagents are brought to a nozzle where the carrier fluid (oil) will shear the water phase into small droplets at high speeds.
Once the encapsulation process is finished, the monodisperse emulsion obtained should look like this:
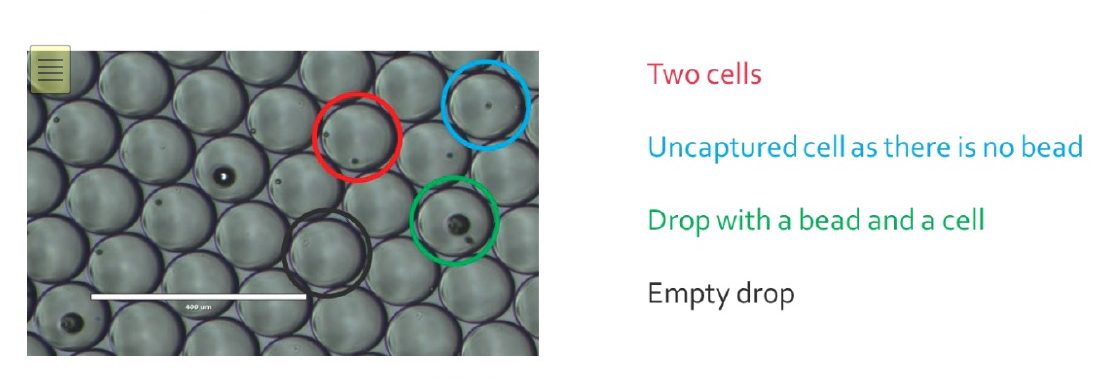
Empty drops will generate noise in the data, as they carry environmental RNA that can bind to beads once droplets get merged.
For drops only containing a bead, most of the time it will not generate more noise as the unused barcodes will be destroyed later in the process.
Cells that were not encapsulated will not be sequenced, as their RNA cannot be captured in drops, but will participate to the noise as the RNA released will bind to random beads.
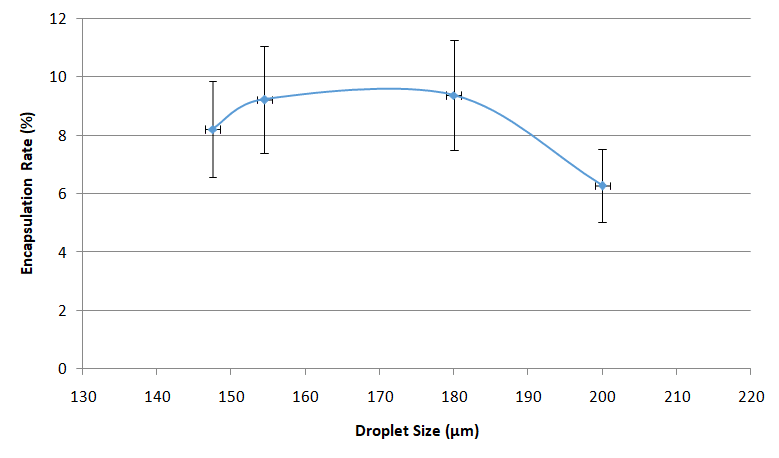
Usually, the Drop-Seq method captures around 5-10% of the input cells.
This is due to the Poisson distribution, basically, this is a balance between having too many empty drops and having drops with two or more cells/beads, that will generate noise and unusable data during sequencing.
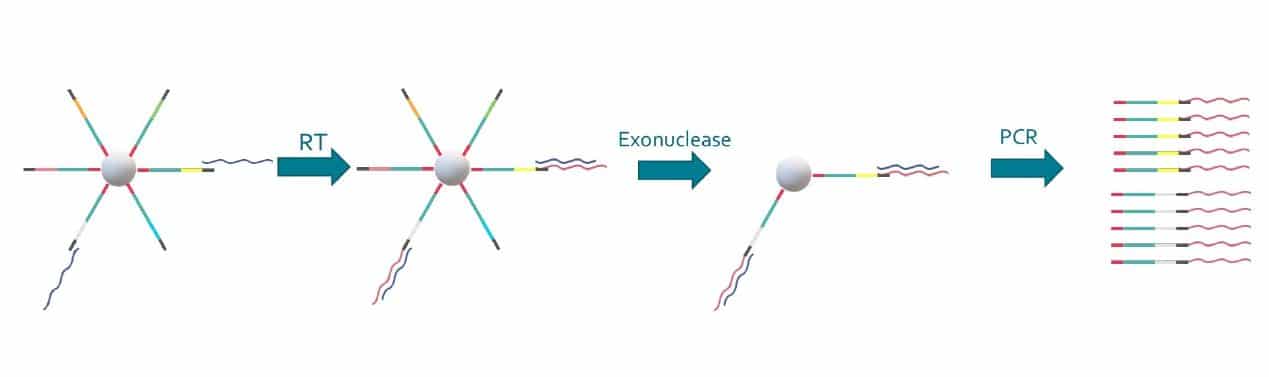
Special buffer components that are contained in the bead buffer will start cell lysis as soon as the drops are formed, and will carry on during droplet production. By the time the run is finished, all the cells will be lysed and RNA captured. This means the emulsion may be broken by merging all the drops and recovering the beads to start the molecular biology steps for sequencing the captured RNA. Reverse transcription (RT) will generate cDNA attached to the barcodes, and an exonuclease will be used to remove unused barcodes.
Promising results from the Drop-Seq protocol
Different genes, one cell?
First, there is the PCR handle, which will allow for the amplification of the generated cDNA molecules at the end of the process, mainly for amplifying the signal by generating more copies.
Second comes the barcoding sequence, which will be the same on each oligo from one bead, but different from one bead to another. All the sequences that contain a given barcode will then be associated to the same cell of origin.
Third comes the UMI, which stands for Unique Molecular Identifier. All the oligos on one bead have a different UMI sequence, so that different molecules of mRNA can be distinguished at the cell level. This allows for one to know how many mRNA of a given sequence have been produced by a cell.
Finally comes the capture sequence, which is usually a poly(T) tail to match the poly(A) tail of the 3’ end of mRNA. If you are only targeting a specific gene panel, you can also use complementary sequences to only capture those.
The power of the Drop-Seq method relies on how the microbeads are constructed: they are coated covalently with oligonucleotides that help capture RNA, barcode it in a way each bead contains unique barcode and diverse unique molecular identifiers to help quantifying captured RNA molecules.

Conclusion
According to Macosko and by following the McCarroll protocol:
“Drop-Seq can prepare 10,000 single-cell libraries for sequencing in 12 hours, for about 6.5 cents per cell representing a >100-fold improvement in both time and cost relative to existing methods”
Using the Fluigent Drop-Seq method with pressure will allow to have same results with more control on droplet size and then better reproducibility.
Fluigent setup can also be automated using OxyGEN software for routine experiment as well.
Expertises & resources
- Expertise videos
MICROFLUIDICS in DROPLET DIGITAL PCR
Read more - 어플리케이션 노트
Encapsulation of multiple emulsions in a single droplet
Read more - 어플리케이션 노트
Droplet Sequencing: Drop-Seq method
Read more - 기술서
Droplet-based Microfluidics – A Complete Guide
Read more - Product presentation videos
DROPLET STARTER package – Make DROPLETS within minutes!
Read more - Support & Tools
Droplet Size Calculator
Read more - 미세유체학에서의 물방울 및 입자 생성
Microfluidic Droplet Production Method
Read more