Microfluidics for vaccines research and development
The WHO estimated global demand for vaccines of 3.5 billion doses in 2018, up approximately 25% compared to 20171. With the demand for current vaccines, the appearance of new viruses such as Ebola and COVID-19 leads to an even more critical demand. A report from IPBES also states that the increasing exploitation of the environment will increase the occurrence of new pandemics2. All this put together raises pressure on vaccine development time and cost.
THE MISSION OF FLUIGENT IS TO DEVELOP SOLUTIONS TO SUPPORT RESEARCHERS AND INDUSTRIALS CONDUCTING MICROFLUIDIC EXPERIMENTS UNDER EXCELLENT CONDITIONS.
Technological advances for vaccine development
First-generation vaccines are based on whole organisms, in either live attenuated or killed forms. Second-generation vaccines consist of protein components derived from the organism. Today, third-generation vaccines are being developed. These nucleic acid vaccines are based on the genetic material of the infectious organism. DNA vaccines are examples of third generation vaccines. Clinical trials for DNA vaccines to prevent HIV are underway.
During the COVID-19 pandemic, the first vaccines to reach clinical trials were based on viral vector and nucleic acid technologies. One of the most promising vaccine candidates was based on mRNA.
Scientific advances are becoming even more critical in the success of fundamental vaccine research, vaccine development, or diagnostics. Microfluidic methods can be used to improve vaccine research and development.
Microfluidics in vaccine research and development
Efficient delivery of mRNA and DNA into target cells in vivo is a major challenge. Today, many vaccines are encapsulated into nanoparticles, specifically lipid nanoparticles (LNP) such as liposomes, allowing for improving efficient delivery. The first COVID-19 vaccines to reach the market are mRNA vaccines encapsulated in LNP.
Size and size distribution are essential properties that determine the clinical successes of the nanocarriers. Recently, improvements have been made with the development of microfluidic production methods, in which LNPs formation occurs within a confined microenvironment. These methods have demonstrated higher control over the physical properties of the end product, particularly in terms of liposome size and size distribution.
Microfluidic application example
Vaccine adjuvant production

In vaccine development, adjuvants are used to amplify the recipient’s specific immune responses against pathogen infection. A new generation of adjuvants is being developed to meet the demands of improved antigen specific responses with less toxicity. For example, Konishi et al. reported the first synthesis of a series of saponins, a vaccine adjuvant, using a microfluidic mixer3.
High throughput screening
Flow cytometry for single single-virus analysis shows high potential in virology, vaccine development, or antiviral research. However, it is hard to perform such analysis as Fluorescence-activated cell sorting (FACS), the standard technique in modern biology, does not allow single-virus measurements due to the small size of viruses.
The Fluigent double emulsion production station is a robust and complete system for producing outstanding monodispersed double emulsion in one single device.
Droplet-based microfluidics combined with next-generation sequencing can allow for screening virus particles in a high throughput manner. For instance, Chaipan et al. developed a droplet-based microfluidic platform to screen single virus particles for optimal antigenic features of vaccine candidates. They demonstrated its use by screening HIV particles4.
The Drop-Seq protocol, is a high throughput method that enables the sequencing of the mRNA from a large number of cells.

Virus detection and analysis
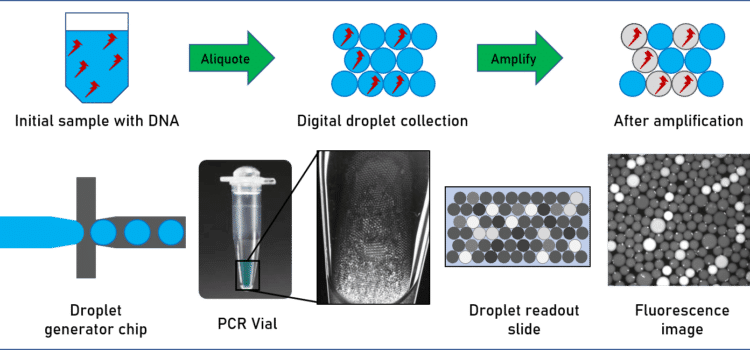
Polymerase chain reaction (PCR) and other PCR related methods such as quantitative PCR (qPCR) amplify and quantify DNA and RNA for subsequent analysis. In contrast to standard PCR, digital PCR (dPCR) divides the sample into subsamples of pico to nanoliter range, allowing for a more reliable collection and sensitive measurement of nucleic acid amounts.
For example, Ahrberg et al. developed an easy-to-use microfluidic dPCR device to amplify and quantify complementary deoxyribonucleic acid (cDNA) samples for the H7N9 influenza5. In another study, researchers from the California Institute of Technology and MIT developed a microfluidic dPCR approach to physically link single bacterial cells harvested from a natural environment with a viral marker gene for examining virus-bacterium interactions in many environments6.
In-Vitro cell analysis
Microfluidics provides unique capabilities to control the cellular microenvironment and present cells with mechanical and biochemical signals in a more physiologically relevant context. Microfluidic chips offer an ideal microenvironment to study molecular- and cellular-scale activities that underlie human organ function and identify new therapeutic targets in vitro. For instance, Markus et al. used microfluidic cell culture chambers to analyze the hosting of varicella-zoster virus in human embryonic stem cell-derived neurons7.

References
- World Health Oganization. Global vaccine market report. 14 p (2019).
- Luthando Dziba, Isabel Sousa Pinto, Judith Fisher, K. T. IPBES Workshop on biodiversity and pandemics. (2020).
- Konishi, N. et al. Synthesis of Bisdesmosidic Oleanolic Acid Saponins via a Glycosylation-Deprotection Sequence under Continuous Microfluidic/Batch Conditions. J. Org. Chem. 82, 6703–6719 (2017).
- Chaipan, C. et al. Single-Virus Droplet Microfluidics for High-Throughput Screening of Neutralizing Epitopes on HIV Particles. Cell Chem. Biol. 24, 751-757.e3 (2017).
- Ahrberg, C. D., Lee, J. M. & Chung, B. G. Microwell Array-based Digital PCR for Influenza Virus Detection. Biochip J. 13, 269–276 (2019).
- Arbel D. Tadmor, Elizabeth A. Ottesen, Jared R. Leadbetter, and R. P. Probing Individual Environmental Bacteria for Viruses by Using Microfluidic Digital PCR. Science (80-. ). 23, 1–7 (2012).
- Markus, A., Lebenthal-Loinger, I., Yang, I. H., Kinchington, P. R. & Goldstein, R. S. An In Vitro Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived Neurons. PLoS Pathog. 11, 1–22 (2015).